Computer Vision and Identifying Patterns in Breast Cancer
Background
In late 2019, I was fortunate to have had the opportunity to lead an outcome-driven machine learning project while at Slalom Consulting with the American Cancer Society in honor of a colleague that had passed away from cancer. As we explored different topics and conducted workshops on areas of collaboration where our skills (computer science, artificial intelligence, epidemiology, population science) might complement each other, we ultimately landed on a use case that was very near and dear to my heart. I hope this high-level overview conveys to any reader the overall process, things to consider, challenges, and opportunities of this type of work.
The Problem
Statistically, we know that 1 in 8 women in the United States alone will be diagnosed with breast cancer in their lifetime. Many of us, myself included, have already lost or will lose family members and people close to us to this complex disease.
We also know that pathologists, highly specialized scientists that train in understanding specific diseases especially from samples (e.g. tissue samples), are high in demand and low in supply. There simply are not enough pathologists to support the increasing demand of cases.
However, with the rise of digital pathology (expected to grow year of year by 13% to 2026 according the Allied Market Research) and the digitization of tissue samples, there are opportunities to augment pathologists and scale the review of tissue samples using machine learning to support them (not replace them) with the consistent, tireless ability of computers. Machines also have the potential to see information at the pixel level in images that cannot be seen by the human eye.
This was what we wanted to research and explore the feasibility of identifying patterns across digital images of breast cancer tumor tissue samples using computer vision.
The Project
The American Cancer Society has a rich history of collecting data on cancer through major cohort studies, called Cancer Prevention Study. These studies track participants throughout their lifetime via longitudinal surveys and provide invaluable insights to us about how cancer develops all the way from lifestyle factors, diagnoses, and now even genomic information. When we began our collaboration, they had just digitized 1,700~ breast cancer tumor tissue samples collected via Cancer Prevention Study II (20 years) from across 847 hospitals across the United States.
We wanted to explore the following questions:
Could machine learning, specifically computer vision and deep learning, be used to identify patterns across all of these images without explicitly being told what to look for (unsupervised)?
Could there be novel patterns that only a machine would be able to see?
Based on these patterns, could we group the images according to shared features? Would these groupings result in logical groupings that could support or accelerate the work of pathologists?
To effectively determine if these questions were the right ones to explore together, and if we wanted to go on this journey together, it was critical to consider:
Is this an appropriate level of scope and in what time frame could this exploration be accomplished?
Is there a values and interest alignment between the two teams?
Is there significant research/social value to answering this question? How will success be measured?
Is there sufficient interdisciplinary representation to conduct a collaboration? It is a common misconception that machine learning projects can be completed with computer scientists alone. The reality is that projects will have better results with the right mix of diverse backgrounds and perspectives.
Is this feasible from a technical perspective? Did we have enough compute and access to GPUs? To address the latter question, we introduced a cloud partner to this project and leveraged the computing resources available there to distribute processing for scale.
The Process
The beginning of these kinds of collaborations typically starts with data, and there are a lot of challenges, even today, when looking at unstructured data like images. The images we worked with for this effort looked like the below.
Hematoxylin and eosin (H&E) stained breast cancer tumor tissue images (tissue samples are not naturally this color - they are stained in a variety of ways).
The dataset we had contained many data quality issues, which is also very normal for any machine learning project. In particular:
Images were collected from over 847 hospitals, meaning the methods used to collect them were not necessarily consistent.
Some of the samples had aged, meaning the colors or staining agents had faded. Others were over-stained and would only add noise to any machine learning model.
The 1,700~ images we had were multidimensional (layered) and were 3-12GB per image. They were also stored in a proprietary image format (.SCN from Leica).
Some samples were physically torn on the glass slide, making them unusable.
To engineer the data into a format that we needed it to be in to run relevant computer vision and deep learning models on top of, we needed to conduct image processing and account for these various data quality issues.
Image Processing
The first step that needed to be done was to convert the images into a standard format. Most machine learning models require image data to be in a standard format to be run on top of them, and .SCN is not one of them. This is a proprietary format specific to Leica and these types of medical images. Unfortunately, there was no easy way to do this at scale (in parallel and pulling out a specific layer of the multiple layers) at the time this project launched, and a custom engineering solution was needed to do this conversion.
Once this hurdle was overcome, we needed a way to address the fading and other data quality issues that included physically torn tissue and hand-drawn sharpie marks on the glass slides that made their way onto the digital images. These erroneous pieces of data are called “artifacts.” We also wanted to avoid wasting compute resources on processing sections of images that were empty space and only focus on parts of an image that had actual tissue on it.
To address the fading and some of the artifacts, we implemented a technique called color normalization. This is essentially where you define specific color ranges - where we take a “golden template” and make sure that, within a certain range, the images we consider have most of their tissue within the same realm of colors, so that “purple” is “purple” across the board. Unnatural colors like that of a black sharpie would be quickly identified and flagged. This technique is not always applied in such studies and may not always be required or beneficial.
For avoiding the processing of white space, we used a formula to identify the ratio of a given image whereby one can determine which areas have a high proportion of white space versus actual tissue in the image.
Now, we realized the images were too large to process (what is called whole-slide image processing) and so yet another technique was combined with all of these others called “tiling.” This is exactly what it sounds like, where one massive image (remember, up to ~10GB per) is broken out into several smaller "tiles.” Doing this makes it easier to eliminate smaller portions (tiles) that have artifacts, as well as those with white space in a way that allows us to break out the compute in a distributed fashion - meaning each tile can be processed at the same time across many machines. This is the most resource intensive part of the process and can be very costly, making it potentially prohibitive to some (more on this later).
Sample of whole slide image “tiles”
Once this entire process is done, the modeling process can be started.
Feature Engineering
Since the objective of this exploratory collaboration was to group images based on patterns a machine sees versus logical groupings a human would use, we needed to leverage what are called “unsupervised” methods. Whereas in “supervised” methods we would tell the computer what a “cat” is and feed it millions of images labeled by humans as “cats,” in unsupervised we must find ways for the machine to do the remaining steps, including feature engineering.
Feature engineering refers to the part of the machine learning process where we would transform raw data or parts of raw data into engineered data based on domain knowledge that will be more effective at predicting what we want. This might mean using finance expertise to use the standard deviation of something instead of a raw number.
There are ways to do this process in an unsupervised fashion. One of these techniques is called an “autoencoder” and is often applied in computer vision and other unstructured data problems.
Sample autoencoder image from Wikipedia
The job of the autoencoder is to take the “tiled” image in this case and reduce it down into a numeric vector representation of the main components and patterns within it while still being able to replicate as much as possible the original input. This was applied to all tiles and covered the question around using “machine determined” patterns, and the output was then fed into a second type of machine learning (specifically, deep learning) model.
Clustering
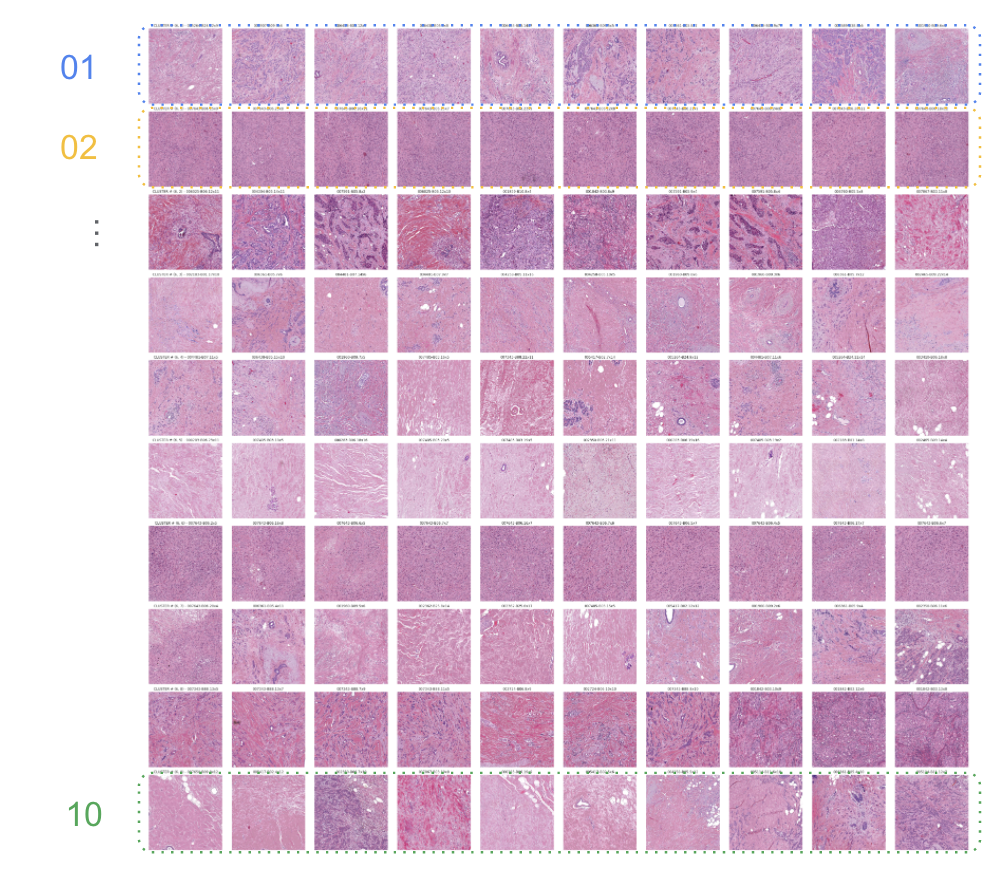
The original hypothesis was not only to see if the machine could identify patterns within the images, but to also group them. To do this, we leveraged multiple rounds of clustering with an algorithm known as K-means. This grouped the results of the previous step into 10 groups as in the image below (representative only):
Representative image of clustering of tiles from Google NEXT slides
Outcomes
With the computer generated clusters grouped by features identified by unsupervised techniques, it was then up to the research groups to conduct further research and determine the novelty of the groupings based on then combining the tiles/tissue samples to the underlying longitudinal survey information from Cancer Prevention Study II. More research on this front can be found in the links below.
There is also a desire to open source this effort and contribute it back to the community, so that others may continue the study or enhance the process.
We understand these types of technological advances have the opportunity to improve the outcomes for many and augment the work of pathologists across the world, but we must also ensure a level of accessibility as they are developed.
Opportunities for the future
The collaboration had a lot of learnings and findings, but there is always room to do more in this space to drive better outcomes for patients. Some of the considerations include:
Reduce computational effort - To make this type of technology more readily available to all areas and patients, the barrier to entry caused by both the compute resources and specialized human labor skills needed must be reduced. The nature of medical images makes them complex and expensive to process at scale.
Increase data distributions of source information - Many organizations are working on increasing the diversity of data collected for studies and this should continue to be an area of focus to ensure everyone is being taken into consideration.
Create more explainability of deep learning models - There are many innovations on this front, but it is still an area of improvement in terms of understanding how some of these models come to specific conclusions. We should also be able to replicate any study out there, which is not always the case.
Increase accessibility/adoption of digital pathology - Not everyone has access to digital pathology, as it often requires high infrastructure costs to adopt on the hardware side (e.g. specific scanners, integrations, etc.). There are many professional organizations pushing this adoption, but this is another area of opportunity since it would unlock the ability for pathologists anywhere to review and discuss cases.
Reference
Allied Market Research. “Report.” 2020.
AACR. “A scalable, cloud-based, unsupervised deep learning system for identification, extraction, and summarization of potentially imperceptible patterns in whole-slide images of breast cancer tissue.” 2019. Link.
WABE. “Using artificial intelligence to fight breast cancer.” 2019. Link.
Images from original public Google NEXT presentation. 2019. Link. Link 2.
American Cancer Society. “About Breast Cancer.” 2021. Link.
American Cancer Society. “History of Cancer Prevention Study.” 2021. Link.
Wikipedia references.